The heart is a mechanical machine that has little room for failure. We still do not understand how the molecules that make up the heart ensure the remarkable reliability of this organ. In this context, our focus is on the following questions: How do the mechanical properties of proteins like titin and cardiac myosin binding protein C (cMyBP-C) emerge into the tissue scale to set the overall mechanics of the myocardium? How is protein mechanics modulated during physiological adaptation or altered in diseases of the heart? How do genetic variants in mechanical proteins cause cardiomyopathy? Our laboratory addresses these questions following multidisciplinary approaches, always trying to develop novel enabling methods and collaborating with outstanding colleagues whenever possible. A main strength of the laboratory is our command of single-molecule biophysics techniques for the mechanical characterization of proteins, including force spectroscopy by atomic force microscopy (AFM) and magnetic tweezers (MT), as well as complementary computational methods. To provide biological context to our results, we also measure mechanics at the level of cardiomyocytes and myocardial preparations and take advantage of our expertise in protein biochemistry and molecular and cell biology. In our pursue of biological contextualization, we have recently expanded the range of mechanical manipulation of proteins to living cells and animals. There is more info about current projects in the lab below.
You can find a list of Jorge’s publications here.
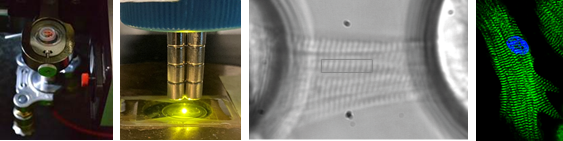
Protein mechanics in vivo
With the aim of moving the field of protein mechanics from correlations to causality, we are committed to developing methods to study protein mechanics in vivo. Many of these tools are based on the use of specific proteases to cease force transduction across mechanical proteins at will. Our efforts are funded by the ERC-Consolidator program (ProtMechanics-Live) and are based on the HaloTag-TEV-titin mouse model, which we reported in Nature Communications 2020, 11, 2060 in collaboration with the group of Wolfgang Linke (U. Münster). The application of this strategy to titin is producing results that were inaccessible to date and that we are currently studying in depth. We have not published our results yet, but if interested, send Jorge a message because we would love to contribute our tools to address relevant questions outside our comfort zone!
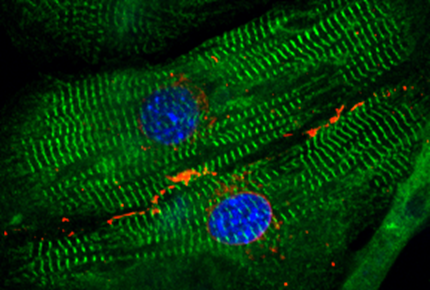
Force-producing structures in cardiomyocytes
Crosstalk between protein mechanics and heart pathophysiology
Within our general interest to understand how protein mechanics contributes to the pathophysiology of the heart, a traditional research line of the lab has been devoted to redox posttranslational modifications targeting cysteines, that are potent modulators of titin mechanics (Cell 2014, 156(6):1235, Nature Communications 2018, 9:185 reviewed in Redox Biology 2019, 21:101074). Recently, we have been able to show that these oxidations target titin in vivo (Redox Biology 2022, 52:102306), which triggers questions about their biological origin and how cardiomyocytes can take advantage of them to modulate mechanical output. Prompted by our results, we are now unraveling the mechanical consequences of other posttranslational modifications that are prevalent in diseases of the heart and may contribute to pathophysiology.
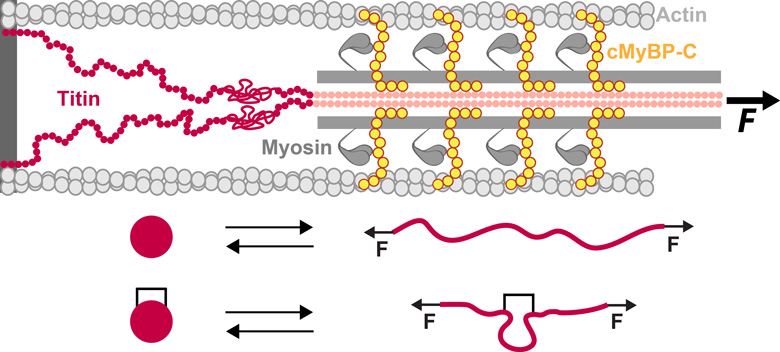
Titin and cMyBP-C are proteins with mechanical roles in the sarcomere.
The unfolding of protein domains under force is affected by
crosslinking posttranslational modifications such as disulfide bonds.
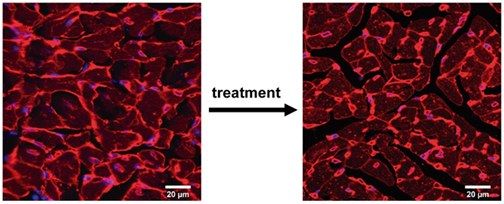
Mechanical proteins and the origin of familial cardiomyopathies
Familial cardiomyopathies are genetic diseases of the heart that account for the majority of cases of sudden cardiac death in young people. They are also the number one cause of heart transplantation worldwide. It turns out that mutations in genes coding for mechanical proteins are the most common cause of cardiomyopathy, although the reasons remain largely unknown. We have recently demonstrated that specific physicochemical features, i.e. reduced hydrophobicity of core residues of protein domains, fingerprint pathogenic variants in titin that cause dilated cardiomyopathy (DCM) (Cell Reports 2023, 42, 113490). We are now investigating why, with the final aim of finding new therapeutic windows of opportunity in this condition. We are also interested in hypertrophic cardiomyopathy (HCM), the most common genetic disease of the heart. Following up on our observation that many pathogenic missense variants in cMyBP-C that cause HCM do not show any classical pathogenic molecular phenotype (J Biol Chem 2021, 297, 100854, ACS Nano 2021, 15, 10203), we have developed new animal models to test novel pathomechanisms and the potential efficacy of therapies for this important group of patients (check our latest preprint BIORXIV/2024/584986).