Difference between revisions of "QuiXoT"
| Line 16: | Line 16: | ||
{{See also|DataGrid information in QuiXoT}} | {{See also|DataGrid information in QuiXoT}} | ||
| − | === Checking an existent QuiXoT analysis === | + | === Part I: Checking an existent QuiXoT analysis === |
==== The QuiXML files ==== | ==== The QuiXML files ==== | ||
| Line 61: | Line 61: | ||
If you have used 18O-labelling, you can check the labelling efficiency, prior to other analyses. Plot ''q_f'' versus ''Xs'' (consult how to create graphs). Since this plot does not differentiate between good and bad quantitations and hence plots together more and less accurate estimations of ''q_f'', it is a good idea to eliminate bad quantitations from the plot by filtering out the data that do not have an arbitrary minimum ''Vs'' value (for instance ''Vs > 30'' in ZoomScan-quantitated spectrum). Labelling efficiency must be above 0.8 for the vast majority of peptides. A cloud of points with ''q_f'' below 0.7 tending to curve towards the right (increasing ''Xs'' values) are indicative of a poorly-labeled experiment. | If you have used 18O-labelling, you can check the labelling efficiency, prior to other analyses. Plot ''q_f'' versus ''Xs'' (consult how to create graphs). Since this plot does not differentiate between good and bad quantitations and hence plots together more and less accurate estimations of ''q_f'', it is a good idea to eliminate bad quantitations from the plot by filtering out the data that do not have an arbitrary minimum ''Vs'' value (for instance ''Vs > 30'' in ZoomScan-quantitated spectrum). Labelling efficiency must be above 0.8 for the vast majority of peptides. A cloud of points with ''q_f'' below 0.7 tending to curve towards the right (increasing ''Xs'' values) are indicative of a poorly-labeled experiment. | ||
| − | === Analysing an experiment from scratch === | + | === Part II: Analysing an experiment from scratch === |
| − | ==== Generating the files === | + | ==== Generating the files ==== |
Generate the QuiXML file containing the list of identified peptides. You can do this in different ways: | Generate the QuiXML file containing the list of identified peptides. You can do this in different ways: | ||
Revision as of 17:06, 12 September 2013
| QuiXoT |
|---|
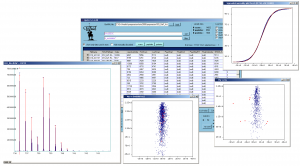 Screenshot of QuiXoT, depicting different spectra and graphs used. |
| Last release: v.1.4.00 |
| Release date: 20th Aug 2013 |
| Download link: [[{{{link}}}]] Source code: QuiXoT at GitHub |
| Licence: Please read Licencing |
| Requirements |
|
QuiXoT is an open source software created for the quantitation and statistical analysis of quantitative proteomics experiments. It has been developed at the Cardiovascular Proteomics Laboratory of Prof Jesús Vázquez, at the Centro Nacional de Investigaciones Cardiovasculares (CNIC), Madrid, Spain.
It has been developed in Visual C#, hence users must install the .NET Framework 2.0 or higher (not necessary for Windows 7 users), which can be downloaded from this link.
Contents
Using QuiXoT
- See also the article: DataGrid information in QuiXoT.
Part I: Checking an existent QuiXoT analysis
The QuiXML files
QuiXoT makes use of a QuiXML files, which is an ad hoc XML format created to manage the three levels of information treated: identification, quantitation and statistical information. To check a list of the different fields used in QuiXML files (i.e., the columns appearing in the main window of QuiXoT), you can check the article DataGrid information in QuiXoT.
If you just want to see an existing QuiXoT analysis, you only need the corresponding QuiXML file. Just drag and drop that file on the main form, and select the quantitation method used, which will depend on the SIL method used (such as 18O, SILAC, etc) and the spectrometre used (sich as high or low resolution).
The binStack folder
The spectra are saved in a folder called binStack, which contains one or more .bfr files and one index.idx file. You do not need this folder if you just want to check the results of a QuiXoT analysis (such as the statistics, identifications or the quantitative information).
However, you will need it if you want to requantitate a spectrum, or see the spectrum itself (for example to compare the theoretical and experimental isotopic envelope, which are respectively in red and blue colours). If this is your case, then you should always have the QuiXML file and its corresponding binStack in the same folder (do not forget to move them together).
As far as the binStack and the QuiXML file are in the same place, you do not need to do anything else to load the spectral information.
The configuration files
In the location where you have copied your version of QuiXoT you will find a conf folder containing the configuration files. It contains three kinds of file:
- the QuantitationMethods.xml file, which contains the parameters of the different methods used. Here it is specified which labelling is associated to a method, or which is the spectrum type that contains the quantitative information (for instance, SILAC quantitation is performed in the full scan if a high resolution spectrometre has been used, but if it is a low resolution machine, then the quantitation is performed in the zoom scan immediately previous to the MS2 scan). Examples of other parametres that can be determined using this file are:
- width: the tolerance for the high resolution peaks
- deltaR: the mass difference between 16O and 18O
- sumSQtolerance_NG, the tolerance accepted for the sum of squares when comparing the theoretical and the experimental spectrum
- the iTRAQ masstags and their corrections
- several xml files which containg information such as the weights of each isotope, the composition of each amino acid or their posttranslational modifications. They also include the correspondence between the different residues and their symbols; for instance, "Y" means "tysorine", while "*" may refer to an oxidation in methionine, or a SILAC label on arginine. Examples of these files are:
- isotopes.xml
- aminoacids.xml
- aminoacids_SILAC.xml
- several xsd files, which are the XML schemas that contain the structure of the QuiXML file depending on each quantitation method. Some examples of these files are
- identifications_schema_18Ohighres.xsd
- identifications_schema_mascot_SILAC.xsd
Checking spectra by weight
Inspect quantifications with low Vs values. Sort the table by Vs and inspect the spectra by using the spectrum button. At very low Vs values you will find completely useless spectra (bad fittings, mixtures, high background, etc). You can choose whether eliminating these spectra from the statistics by marking them with numLabel1 = 0, or filtering by a minimum Vs value (for instance Vs > 3). Non-quantifiable peptides (i.e. peptides not containing basic N-terminal residues in 18O-labeling or in SILAC) must also be excluded when calculating variances or performing the statistics.
Labelling efficiency for 18O labelling
If you have used 18O-labelling, you can check the labelling efficiency, prior to other analyses. Plot q_f versus Xs (consult how to create graphs). Since this plot does not differentiate between good and bad quantitations and hence plots together more and less accurate estimations of q_f, it is a good idea to eliminate bad quantitations from the plot by filtering out the data that do not have an arbitrary minimum Vs value (for instance Vs > 30 in ZoomScan-quantitated spectrum). Labelling efficiency must be above 0.8 for the vast majority of peptides. A cloud of points with q_f below 0.7 tending to curve towards the right (increasing Xs values) are indicative of a poorly-labeled experiment.
Part II: Analysing an experiment from scratch
Generating the files
Generate the QuiXML file containing the list of identified peptides. You can do this in different ways:
- if you have identified using SEQUEST (which includes Proteome Discoverer), you may use pRatio from either the .msf files (or the .srf files, if you use an older version of SEQUEST).
- if you have identified using Mascot, you may convert the .dat results file using MascotToQuiXML
- if you have used another program, you will need to convert your data into a tab separated text file, and then parse the resulting table using CSVToQuiXML
You will also need the binStack folder, containing the binary files with the spectral information. It can be generated in different ways:
- if you used Thermo RAW files, you can convert the spectra using RawToBinStack
- if you used Mascot generic files (mgf), you can convert the spectra using mgfToBinStack
- if your spectra are stored in a different file format, you should use an external converter to get a mgf file, and the use mgfToBinStack
Performing the first statistics
Introduce an initial set of statical parameters (k and variances) for the null-hypothesis model by using the change_values link. You can find a list of typical values for these parameters. Make an initial estimation of variances by pressing the var calc button. At this step you will have to tell QuiXoT which columns are going to be used as Xs and Vs (this is useful for multichannel labelling approaches such as, for instance, iTRAQ data, which contains several Xs and Vs values depending on the labels that are to be compared). Accept the newly calculated variances and perform the statistical analysis by pressing the stats button.
Inspecting spectra and peptides

Inspect the presence of outliers at the scan and peptide levels by using the graphs button and setting Ws (or Wp) as X, vs Xs (or Xp) as Y, to check whether these data are influencing variance calculation. Sort out the data by FDRs (or FDRp) and check the rows having low FDR values (below 0.05 they are statistically considered as outliers). Typically a negligible proportion of outliers may be found (less than 1% of total); this is normal. However, if the number of outliers is too high, it may be indicative of quantification artefacts and/or problems in the labelling protocol.
Common artefacts at the scan level are rare and may be produced by
- a) problems in mass calibration (spectra cannot be fitted to the theoretical mass envelope)
- b) excesive noise and/or fluctuations in the detector
- c) inadequate fitting parameters in the configuration files.
Common artefacts at the peptide level are, however, much more frequent when peptides are post-digestion labelled (which does not include SILAC). They include:
- a) incomplete digestion of one of the samples (this may be easily checked by selecting peptide subpopulations using the st_PartialDig field and the filter tool in Vs versus Xp plots)
- b) non homogeneous methionine oxidation in the samples to be compared (this may be easily checked by filtering out by the st_Meth field)
- c) partially labelled peptides (with 18O-labelling this is indicated by q_f)
If any of these artefacts are encountered, outliers should be eliminated from variance calculation or statistics by using the filter tool.
A further inspection of proteins showing significant expression changes (low FDRq values) is recommendable at this step, since keratins and other external contaminants like trypsin may not be well-balanced in the two samples and introduce an artefactual variance at the protein level. Eliminate all the quantifications related to these contaminants from the statistics by applying an appropriate filter (consult applying filters to the data).





