Nucleic Acids Research: A CNIC team has created an innovative tool for the reliable and efficient study of gene function
iSuRe-HadCre promises to become an essential tool for biomedical research using mouse models to modify and understand gene function
A team of scientists at the Centro Nacional de Investigaciones Cardiovasculares (CNIC) led by Rui Benedito has generated a novel genetic tool, called iSuRe-HadCre, that enables the induction of precise genetic alterations in whole tissues or in individual cells with high efficiency and reliability. This new technology is set to become an important tool in biomedical research that uses mouse models to modify and understand gene function.
For decades, the gold standard method for analyzing gene function has been conditional Cre-recombinase genetics, which offers precise control over genetic alterations or gene expression. Despite the emergence of new gene targeting technologies like CRISPR/Cas9, the Cre-Lox system in mouse models remains unparalleled in its efficiency, precision, and broad applicability in biomedical research. This method allows researchers to manipulate gene expression temporally and spatially, enabling the study of gene function in specific tissues and in specific biological or disease contexts.
However, a major drawback of the Cre-Lox system is its unavoidable variability in the efficiency and temporal control of Cre-mediated recombination depending on the specific transgenes and floxed alleles used. This variability requires the use of diverse and costly genetic and molecular controls to confirm that each Cre-dependent conditional genetic experiment has worked as intended.
To overcome these limitations, the CNIC team led by Rui Benedito previously developed the iSuRe-Cre technology, a transgenic mouse model that linked permanent Cre activity to the expression of a fluorescent reporter. This overcame the problem of false positives and the need for costly additional checks and controls to validate appropriate modification of the targeted genes. Nevertheless, this first-generation iSuRe-Cre allele still had limitations, including sporadic leakiness in some cell types, relatively low sensitivity to Cre activity, and the potential for toxicity in cells with high and permanent Cre expression.
In the new study, the CNIC team developed a new genetic tool called iSuRe-HadCre that overcomes these limitations. “This tool uses a novel inducible dual-recombinase genetic cascade that ensures expression of a fluorescent reporter in all cells that previously had, but no longer have, high Cre activity,” said first author Irene García González.
García González explained that “this transient expression of Cre is sufficient to effectively delete all genes flanked by loxP while avoiding toxicity caused by the permanent expression of Cre. In addition, the new system is not leaky in any cell type and is much more sensitive to induction by CreERT2 and its ligand tamoxifen.”
Rui Benedito predicts that “these characteristics and the superior performance of iSuRe-HadCre will make it an indispensable tool for all laboratories conducting conditional genetic studies using mouse models.”
Benedito added that iSuRe-HadCre “will be particularly valuable for imaging studies, single-cell genetic analysis, and genetic epistasis studies requiring the simultaneous modification of several genes with high efficiency and reliability.”
The study was funded by the Ministerio de Ciencia e Innovación, "la Caixa" Foundation, the European Research Council, the Leducq Foundation, the Knut and Alice Wallenberg Foundation, and the Göran Gustafsson Foundation.
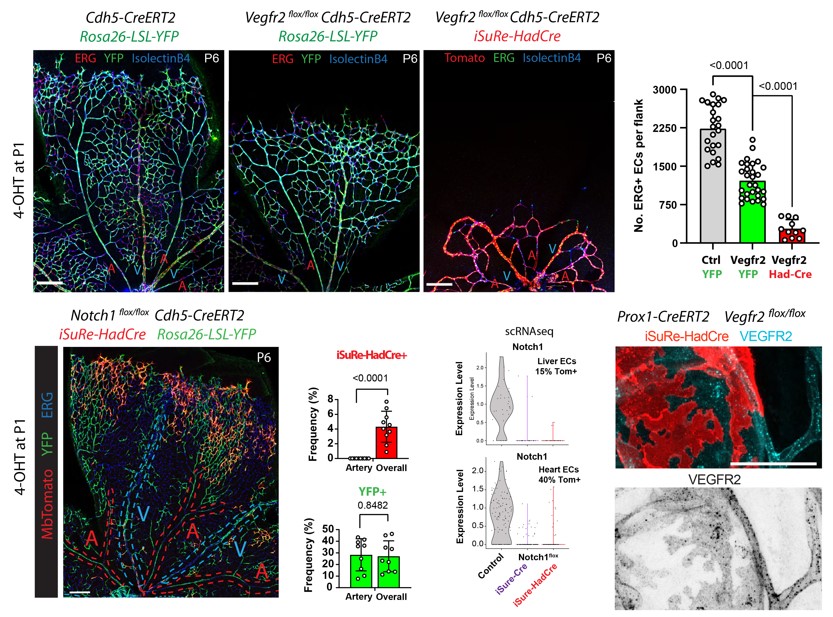
Figure: iSuRe-HadCre is a new tool for effective conditional genetics in the mouse. Images show the comparison between the use of a standard Cre-reporter (Rosa26-LSL-YFP) and the new iSuRe-HadCre allele. Only iSuRe-HadCre ensures effective genetic deletions (Vegfr2 or Notch1) in tissues or single cells.











