Difference between revisions of "Sorting data in QuiXoT"
(creation) |
(No difference)
|
Revision as of 15:57, 13 September 2013
Sorting data
There are two ways of sorting the data in the table:
- Quick sorting: click on the header of the parameter column. An arrow will appear and the data will be sorted in ascending order (alphabetical or numerical depending on the parameter). The order may be reversed by pressing again on the column header. The order (ascending or descending) is indicated by the arrow.
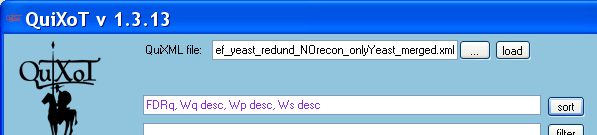
- Multiple sorting: the data may be sorted by several criteria by using the sort field. The command is a list of parameters separated by commas. Sorting is in ascending order by default; to sort in descending order use the DESC command after the name of the parameter. The order of the parameters indicates sorting preference. Once the command is written, press the sort button. If the command is incorrect, no sort will be performed and the command will be highlighted in red.
Example
- The following command
FDRq, Wq desc, Wp desc, Ws desc
- is very useful to analyse significant expression changes. This command will sort out the proteins according to increasing FDRq values (lowest ones in the top of the table), but those with same FDRq and higher Wq will appear first; all scans and peptides corresponding to each protein will appear together, ordered by peptide sequence (higher Wp values first) and scan (higher Ws values first).