Difference between revisions of "Fundamental workflow"
(Created page with "thumb|450px|Schema of the fundamental workflow. The SanXoT workflow consisting of three steps: # '''scan-to-peptide integration''': pep...") |
(No difference)
|
Latest revision as of 11:37, 10 August 2018
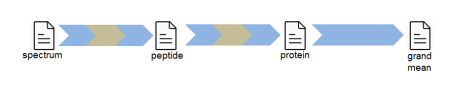
The SanXoT workflow consisting of three steps:
- scan-to-peptide integration: peptide-level quantitative information is obtained by taking into account the information of all the scans where the same peptide is identified. Scan outliers are those that quantify in a statistically significant way compared to other scans pointing to the same peptide (since it is assumed that scans where the peptide is the same, all scans should quantify equally).
- peptide-to-protein integration: protein-level quantitative information is obtained using data of all the peptides that have been identified and quantified for a certain protein. Peptide outliers are those that quantify differently from other peptides pointing to the same protein in the relations file (this might happen especially in non-unique peptides, that can be present in different proteins).
- protein-to-all integration: to compare proteins between them, so proteins having statistically significant changes of expression are identified.
Each integration is performed by the GIA (the Generic Integration Algorithm[1]) In each of these steps, the associated level variance can be calculated, giving detailed information about the experiment[2].
There are many possible variations of this workflow. For example, the last step (protein-to-all) can be removed to perform a peptide-level systems biology, or the peptide-to-protein integration can be modified to account for post-translational modifications[3], or extra levels can be added prior to the scan level (for example to integrate different features present in the same spectrum, as in the case of NeuCode[4]).
See also
- The fundamental workflow at Exploring SanXoT features
- Unit test of the fundamental workflow
- General design of SanXoT
References
- ↑ Garcia-Marques, F., et al., A Novel Systems-Biology Algorithm for the Analysis of Coordinated Protein Responses Using Quantitative Proteomics. Mol Cell Proteomics, 2016. 15(5): p. 1740-60.
- ↑ Bagwan, N., et al. Comprehensive Quantification of the Modified Proteome Reveals Oxidative Heart Damage in Mitochondrial Heteroplasmy. Cell Reports, 2018
- ↑ Herbert, A., et al., Neutron-encoded mass signatures for multiplexed proteome quantification. Nature Methods, 2013