Difference between revisions of "QuiXoT"
| Line 36: | Line 36: | ||
In the location where you have copied your version of QuiXoT you will find a '''''conf'' folder''' containing the configuration files. It contains three kinds of file: | In the location where you have copied your version of QuiXoT you will find a '''''conf'' folder''' containing the configuration files. It contains three kinds of file: | ||
| − | * the '''QuantitationMethods.xml''' file, which contains the parameters of the different methods used, | + | * the '''QuantitationMethods.xml''' file, which contains the parameters of the different methods used. Here it is specified which labelling is associated to a method, or which is the spectrum type that contains the quantitative information (for instance, SILAC quantitation is performed in the full scan if a high resolution spectrometre has been used, but if it is a low resolution machine, then the quantitation is performed in the zoom scan immediately previous to the MS2 scan). |
| − | * several xml files which containg information such as the weights of each isotope, | + | * several '''xml files''' which containg information such as the weights of each isotope, the composition of each amino acid or their posttranslational modifications. They also include the correspondence between the different residues and their symbols; for instance, "Y" means "tysorine", while "*" may refer to an oxidation in methionine, or a SILAC label on arginine. Examples of these files are: |
:* isotopes.xml | :* isotopes.xml | ||
:* aminoacids.xml | :* aminoacids.xml | ||
| − | * several xsd files, which | + | :* aminoacids_SILAC.xml |
| + | * several '''xsd files''', which are the XML schemas that contain the structure of the QuiXML file depending on each quantitation method. Some examples of these files are | ||
:* identifications_schema_18Ohighres.xsd | :* identifications_schema_18Ohighres.xsd | ||
:* identifications_schema_mascot_SILAC.xsd | :* identifications_schema_mascot_SILAC.xsd | ||
[[Category:QuiXoT]] | [[Category:QuiXoT]] | ||
Revision as of 16:15, 12 September 2013
| QuiXoT |
|---|
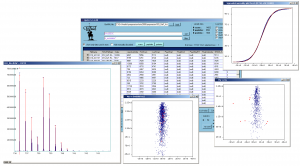 Screenshot of QuiXoT, depicting different spectra and graphs used. |
| Last release: v.1.4.00 |
| Release date: 20th Aug 2013 |
| Download link: [[{{{link}}}]] Source code: QuiXoT at GitHub |
| Licence: Please read Licencing |
| Requirements |
|
QuiXoT is an open source software created for the quantitation and statistical analysis of quantitative proteomics experiments. It has been developed at the Cardiovascular Proteomics Laboratory of Prof Jesús Vázquez, at the Centro Nacional de Investigaciones Cardiovasculares (CNIC), Madrid, Spain.
It has been developed in Visual C#, hence users must install the .NET Framework 2.0 or higher (not necessary for Windows 7 users), which can be downloaded from this link.
Contents
Using QuiXoT
- See also the article: DataGrid information in QuiXoT.
Checking an existent QuiXoT analysis
The QuiXML files
QuiXoT makes use of a QuiXML files, which is an ad hoc XML format created to manage the three levels of information treated: identification, quantitation and statistical information. To check a list of the different fields used in QuiXML files (i.e., the columns appearing in the main window of QuiXoT), you can check the article DataGrid information in QuiXoT.
If you just want to see an existing QuiXoT analysis, you only need the corresponding QuiXML file. Just drag and drop that file on the main form, and select the quantitation method used, which will depend on the SIL method used (such as 18O, SILAC, etc) and the spectrometre used (sich as high or low resolution).
The binStack folder
The spectra are saved in a folder called binStack, which contains one or more .bfr files and one index.idx file. You do not need this folder if you just want to check the results of a QuiXoT analysis (such as the statistics, identifications or the quantitative information).
However, you will need it if you want to requantitate a spectrum, or see the spectrum itself (for example to compare the theoretical and experimental isotopic envelope, which are respectively in red and blue colours). If this is your case, then you should always have the QuiXML file and its corresponding binStack in the same folder (do not forget to move them together).
The configuration files
In the location where you have copied your version of QuiXoT you will find a conf folder containing the configuration files. It contains three kinds of file:
- the QuantitationMethods.xml file, which contains the parameters of the different methods used. Here it is specified which labelling is associated to a method, or which is the spectrum type that contains the quantitative information (for instance, SILAC quantitation is performed in the full scan if a high resolution spectrometre has been used, but if it is a low resolution machine, then the quantitation is performed in the zoom scan immediately previous to the MS2 scan).
- several xml files which containg information such as the weights of each isotope, the composition of each amino acid or their posttranslational modifications. They also include the correspondence between the different residues and their symbols; for instance, "Y" means "tysorine", while "*" may refer to an oxidation in methionine, or a SILAC label on arginine. Examples of these files are:
- isotopes.xml
- aminoacids.xml
- aminoacids_SILAC.xml
- several xsd files, which are the XML schemas that contain the structure of the QuiXML file depending on each quantitation method. Some examples of these files are
- identifications_schema_18Ohighres.xsd
- identifications_schema_mascot_SILAC.xsd





